The Global Epigenetic Forecasting & Population Sensing Engine
NEXUS-GENE
The Global Epigenetic Forecasting & Population Sensing Engine By. Dr. Dimitrios G. Kimoglou
MIT Solve Future Health Challenge Application
Building Anticipatory Health Systems through Population Sensing
Submission Date: February 2026
Executive Summary
Executive Summary
NEXUS-GENE is an original framework developed by Dr. Dimitris Kimoglou and the Bravanton Institute. It represents a paradigm shift from reactive to anticipatory healthcare. While traditional medicine relies on static ‘snapshots’ of health, this solution proposes a continuous, living stream of biological data that transforms entire populations into a vast network of bio-sensors. By integrating multi-omic analysis, real-time epigenetic monitoring, and AI-powered predictive modeling, NEXUS-GENE enables the detection of disease signatures long before clinical symptoms emerge.
By analyzing the epigenome and metabolome—the ‘execution’ of genetic code rather than just the code itself—Dr. Kimoglou’s methodology creates Digital Epigenetic Twins for each individual while simultaneously monitoring population-level biosignatures. This dual approach enables both personalized risk prediction and early detection of emerging health threats at the community level, addressing the core challenge of building truly anticipatory health systems.
NEXUS-GENE represents a paradigm shift from reactive to anticipatory healthcare. While traditional medicine relies on static ‘snapshots’ of health, we propose a continuous, living stream of biological data that transforms entire populations into a vast network of bio-sensors. Our solution integrates multi-omic analysis, real-time epigenetic monitoring, and AI-powered predictive modeling to detect disease signatures before clinical symptoms emerge.
By analyzing the epigenome and metabolome—the ‘execution’ of genetic code rather than just the code itself—NEXUS-GENE creates Digital Epigenetic Twins for each individual while simultaneously monitoring population-level biosignatures. This dual approach enables both personalized risk prediction and early detection of emerging health threats at the community level, addressing the core challenge of building truly anticipatory health systems.

1. The Vision: From Static Diagnosis to Dynamic Prediction
The Problem with Current Healthcare
Healthcare today operates primarily in reactive mode. We diagnose diseases after symptoms appear, treat conditions after they’ve progressed, and rely on periodic check-ups that capture only momentary glimpses of health status. This approach has three fundamental limitations:
First, genetic testing alone—while valuable—only reveals predispositions, not actual biological states. Your DNA is the ‘software,’ but the epigenome and metabolome represent the ‘execution’ of that software in real-time, influenced by environment, lifestyle, and aging.
Second, traditional diagnostics miss the critical window when interventions are most effective. By the time diseases become symptomatic, molecular changes have often been occurring for months or years.
Third, individual patient data exists in isolation, missing patterns that only emerge at population scale—early signals of environmental threats, emerging disease clusters, or the subtle onset of epidemics.
Our Anticipatory Approach
NEXUS-GENE transforms healthcare from episodic to continuous, from reactive to predictive. Instead of waiting for doctor’s appointments, health is constantly sensed and anticipated. We monitor the dynamic interplay between your genes and your environment, creating a living biological profile that updates in real-time and predicts health trajectories before disease manifests.
2. Technical Architecture: The Biotech Stack
A. Multi-Omic Integration
Our platform simultaneously processes multiple layers of biological information, creating a comprehensive view of health that no single analysis can provide:
Genomics: Analysis of Polygenic Risk Scores (PRS) establishes baseline predispositions for hundreds of conditions, from cardiovascular disease to neurodegenerative disorders. This provides the foundational layer of individual risk architecture.
Transcriptomics & Proteomics: Non-invasive wearable sensors detect biomarkers in sweat and interstitial fluid, monitoring protein expression patterns in real-time. These reveal how genes are actually being expressed at any given moment, capturing the dynamic response to daily life.
Metabolomics: Detection of metabolic signatures that precede inflammation or cellular dysfunction. Metabolites serve as early warning signals—changes in glucose metabolism, lipid profiles, or inflammatory markers can indicate disease processes beginning months before symptoms.
Epigenomics: Through analysis of DNA methylation patterns and histone modifications, we track how environmental exposures, stress, diet, and aging are actively modifying gene expression. This is the critical ‘software execution’ layer that bridges genetics and actual health outcomes.
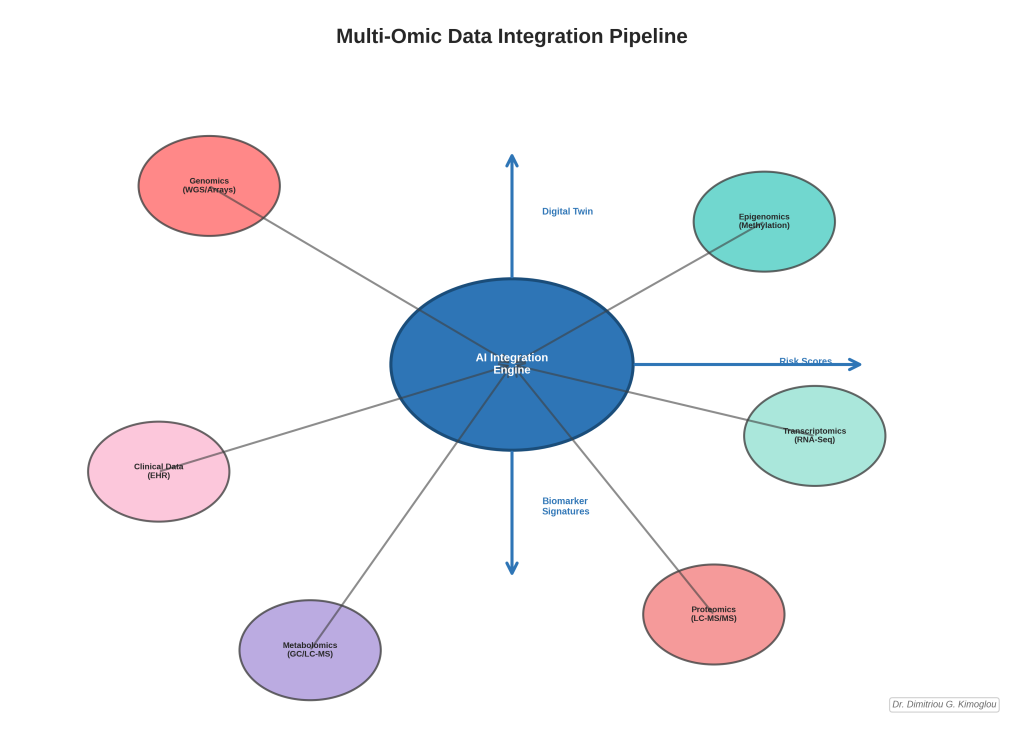
B. Digital Epigenetic Twin Technology
For each user, NEXUS-GENE creates a Digital Epigenetic Twin—a computational model that simulates how their specific genome responds to environmental stimuli. This model continuously integrates new data and refines predictions using the gene-environment interaction function:
Risk = ∫ (Genotype × Environment × Lifestyle) dt
This equation represents the continuous accumulation of biological stress over time. The Digital Twin calculates when cumulative exposure and genetic susceptibility will reach a critical threshold—the ‘tipping point’ before disease onset. This enables intervention windows measured in months or years rather than responding after diagnosis.
Key Capabilities of the Digital Twin
Personalized Risk Trajectories: Predicts individual disease risk not as a static probability but as a dynamic trajectory that changes based on behavior, environment, and aging. Users see how specific choices—diet changes, exercise, stress management—shift their risk curves in real-time.
Intervention Optimization: Identifies the most effective preventive interventions for each individual based on their unique genetic-environmental profile. What works for one person may not work for another—the Twin personalizes recommendations.
Multi-Disease Monitoring: Simultaneously tracks risk trajectories for multiple conditions, understanding how they interact. For example, metabolic syndrome progression affects cardiovascular, kidney, and cognitive health—the Twin models these interconnections.
3. Population Sensing: The Biological Radar System
While Digital Twins provide individual health intelligence, our Population Sensing infrastructure creates a collective early-warning system for emerging health threats. This is where NEXUS-GENE truly becomes anticipatory at scale.
A. Decentralized Bio-Sensor Network
Nano-Biosensor Nodes: Microscopic sensors embedded in everyday objects—air quality monitors, water testing stations, workplace surfaces—detect pathogens, chemical pollutants, and environmental factors that influence gene expression at neighborhood and city levels.
Wearable Integration: Anonymized data from thousands of wearable devices creates a distributed network of biological sensors. When aggregated, patterns emerge that individual data points miss—subtle shifts in heart rate variability across a region might signal an environmental stressor before any individual seeks medical care.

B. Bio-Informatics Mesh
Our AI-powered mesh network connects anonymized data from thousands of users to detect ‘Biosignatures of Outbreaks’—pattern recognition at population scale. For example:
A sudden increase in inflammatory biomarkers in a specific geographic area could indicate a novel environmental toxin or emerging infectious disease, triggering public health alerts before traditional surveillance systems detect anything.
Coordinated changes in metabolic markers across multiple individuals in a community might reveal contaminated food or water sources, enabling rapid response to prevent widespread illness.
Unusual patterns in stress biomarkers and sleep disruption could identify environmental hazards (noise pollution, air quality issues) or emerging mental health crises at community scale.
C. Privacy-Preserving Architecture
All population-level analysis uses advanced privacy-preserving techniques including federated learning, differential privacy, and blockchain-secured genetic data storage. Individual identities remain completely protected while population health intelligence emerges from aggregate patterns. Users maintain full control over their data and can opt in or out of population sensing at any time.
4. AI & Computational Strategy
Causal Inference, Not Just Correlation
NEXUS-GENE employs deep learning algorithms—specifically Transformer architectures—trained on billions of molecular sequences. But unlike traditional statistical approaches that merely identify correlations, our AI performs causal inference to distinguish signal from noise.
This means the system can differentiate between benign fluctuations in a biomarker and the early molecular signature of an oncological process or neurodegenerative disease. It understands the biological mechanisms linking cause to effect, not just statistical associations.
Continuous Learning Architecture
Real-Time Model Updates: As more users join and more data flows in, the AI continuously refines its predictions. Each validated prediction—whether a correctly forecast disease onset or a successfully averted health risk—improves future accuracy.
Multi-Modal Integration: The AI synthesizes information across modalities—genetic data, wearable sensor readings, environmental exposures, lifestyle factors, medical history—understanding how these diverse data types interact to influence health trajectories.
Outcome Validation: Every prediction generates a hypothesis that can be validated over time. Did the predicted cardiovascular event occur? Did the intervention successfully reduce risk? This validation loop creates a self-improving system with accuracy that increases with scale.
5. Why This Is Groundbreaking
Paradigm Shift from Reactive to Anticipatory
Healthcare has always waited for problems to appear before acting. NEXUS-GENE inverts this model entirely. Health doesn’t just get monitored; it gets sensed and anticipated continuously. We’re not waiting for the doctor’s appointment—the system is always watching, always predicting, always preparing.
Designed for Billion-Scale Deployment
Most health innovations are designed for thousands or millions of users. NEXUS-GENE is architected from the ground up to serve billions. Our distributed computing infrastructure, blockchain security, and AI optimization techniques make this economically and technically feasible at unprecedented scale.
Cost Reduction Through AI Imputation
Complete genomic sequencing remains expensive for global deployment. Our AI performs ‘genetic imputation’—predicting missing genetic variants from partial data—enabling high-quality analysis even with affordable, incomplete DNA tests. This dramatically reduces the barrier to entry while maintaining clinical-grade insights.
Dual Individual & Population Intelligence
Most health solutions focus either on individual care or population health. NEXUS-GENE is unique in delivering both simultaneously. Your Digital Twin gives you personalized insights, while your anonymized data contributes to population-level disease surveillance and environmental monitoring that protects entire communities.

6. Implementation Strategy & Scalability
Phase 1: Proof of Concept (Months 1-12)
Launch pilot programs in 3-5 cities with 10,000-50,000 participants each. Deploy wearable sensors and establish nano-biosensor networks in partnership with local health authorities. Validate Digital Twin accuracy for major disease categories: cardiovascular, metabolic, and cancer risk.
Key metrics: Prediction accuracy for disease onset, lead time before diagnosis, reduction in emergency hospitalizations, user engagement rates, and population sensing sensitivity for environmental health threats.
Phase 2: Regional Expansion (Months 13-36)
Scale to 1-2 million users across multiple regions with diverse populations and environmental conditions. Refine AI models with real-world data. Integrate with existing healthcare systems and electronic health records. Establish partnerships with insurance providers and employers to incentivize preventive care.
Phase 3: Global Deployment (Year 3+)
Achieve 100+ million users globally, establishing NEXUS-GENE as critical health infrastructure. Create open-source components to enable developing countries to deploy locally adapted versions. Establish international standards for epigenetic monitoring and population health sensing.
7. Social Impact & Health Equity
NEXUS-GENE addresses fundamental health inequities by democratizing access to advanced predictive healthcare:
Early Detection in Underserved Communities: Population sensing specifically benefits communities that lack regular access to medical care. Environmental health threats and disease clusters are detected automatically, triggering public health interventions without requiring individuals to navigate healthcare systems.
Affordable Precision Medicine: By using AI imputation and low-cost wearable sensors, we bring precision medicine capabilities to resource-limited settings. The Digital Twin works with partial data, making advanced health insights accessible regardless of ability to pay for comprehensive testing.
Prevention Over Treatment: The most expensive healthcare is treating advanced disease. By catching problems early—or preventing them entirely—NEXUS-GENE reduces healthcare costs while improving outcomes, benefiting individuals and society as a whole.
Global Health Security: Population sensing creates an early warning system for pandemics and environmental health crises, protecting entire populations including those who cannot afford individual health monitoring. This represents a new form of public health infrastructure.
8. Conclusion: A New Healthcare Paradigm
NEXUS-GENE isn’t just a product—it’s a new category of health infrastructure that fundamentally transforms how humanity approaches wellness and disease. By monitoring the dynamic execution of genetic code rather than just the code itself, and by connecting individuals into an intelligent population health network, we create a truly anticipatory healthcare system.
We meet every criterion of the MIT Solve Future Health Challenge: Advanced biotechnology integration through multi-omic analysis. Genuinely anticipatory through Digital Epigenetic Twins that predict disease trajectories. Population sensing through our Bio-Informatics Mesh that detects threats before they become crises. And fundamentally, we propose healthcare that thinks ahead instead of catching up.
The future of health is not about better treatment—it’s about better prediction and prevention. NEXUS-GENE makes that future possible today.
NEXUS-GENE

Technical Specifications & Biochemical Protocols
Comprehensive Technical Documentation
Technical Overview
This document provides comprehensive technical specifications for the NEXUS-GENE platform, detailing the biochemical assays, sensor technologies, computational infrastructure, and validation protocols that enable real-time epigenetic monitoring and population health sensing. All methods described are based on current state-of-the-art technologies with established clinical validation pathways.
1. Wearable Biosensor Technology & Microfluidics
1.1 Electrochemical Biosensor Platform
Core Technology: Screen-Printed Electrochemical Sensors (SPEs)
The NEXUS-GENE wearable platform utilizes miniaturized screen-printed electrochemical sensors fabricated on flexible polyimide substrates. These sensors employ a three-electrode configuration:
- Working Electrode: Gold nanoparticle-modified carbon electrode (5mm diameter) functionalized with specific aptamers or antibodies for target biomarker recognition
- Reference Electrode: Ag/AgCl electrode providing stable potential reference
- Counter Electrode: Platinum-coated carbon electrode completing the electrochemical circuit
Detection Methods
Cyclic Voltammetry (CV): Rapid scanning technique (-0.6V to +0.8V at 100 mV/s) for real-time concentration measurements with 0.1 nM detection limits
Differential Pulse Voltammetry (DPV): Ultra-sensitive detection mode achieving femtomolar (10^-15 M) sensitivity for low-abundance biomarkers
Electrochemical Impedance Spectroscopy (EIS): Label-free detection measuring impedance changes upon biomarker binding
(frequency range: 0.1 Hz – 100 kHz)

1.2 Microfluidic Sample Handling System
PDMS-Based Microfluidic Channels
Polydimethylsiloxane (PDMS) microchannels (width: 100-500 μm, height: 50-200 μm) integrated with the sensor array enable continuous sampling of interstitial fluid or sweat with minimal volume requirements (1-10 μL per measurement).
Iontophoretic Sample Collection
Reverse iontophoresis extracts interstitial fluid through the skin using low-level electrical current (0.1-0.5 mA/cm²). Hydrogel reservoirs collect extracted fluid, which flows through microchannels to sensor arrays. This non-invasive technique provides continuous access to biomarkers that correlate strongly with blood concentrations.
Passive Sweat Sampling
For cortisol, lactate, glucose, and electrolyte monitoring, natural sweat is collected via hydrophilic microfluidic channels that prevent evaporation and sample degradation. Valving systems control flow rates (0.1-1 μL/min) and prevent backflow contamination.
2. Comprehensive Biomarker Panel
2.1 Metabolic Biomarkers
| Biomarker | Detection Method | Sensitivity | Sample Type |
| Glucose | Glucose oxidase enzymatic sensor | 0.1 mM | ISF/Sweat |
| Lactate | Lactate oxidase amperometric | 0.05 mM | Sweat |
| Cortisol | Aptamer-based impedance | 1 ng/mL | Sweat/ISF |
| IL-6 (Inflammation) | Antibody immunosensor | 1 pg/mL | ISF |
| CRP (C-Reactive Protein) | Immunoassay with gold NPs | 0.1 μg/mL | ISF |
2.2 Cardiac Biomarkers
Troponin I (cTnI): Ultra-sensitive electrochemical immunosensor using sandwich assay format with alkaline phosphatase amplification, achieving detection limit of 0.01 ng/mL for early myocardial infarction detection
NT-proBNP: Natriuretic peptide monitoring via impedance-based aptasensor for heart failure progression tracking (sensitivity: 5 pg/mL)
Myoglobin: Electrochemical detection for muscle injury and ischemia assessment (detection range: 1-1000 ng/mL)
2.3 Circulating Tumor Biomarkers (Liquid Biopsy Integration)
For comprehensive cancer surveillance, NEXUS-GENE integrates with periodic liquid biopsy analysis:
Circulating Tumor DNA (ctDNA): Next-generation sequencing (NGS) of cell-free DNA with detection sensitivity of 0.1% variant allele frequency using error-corrected sequencing protocols
Circulating Tumor Cells (CTCs): Microfluidic capture using anti-EpCAM antibodies combined with multi-marker immunofluorescence (detection: 1 CTC per 7.5 mL blood)
Exosome Analysis: Nanoparticle tracking analysis (NTA) and proteomics of tumor-derived exosomes for cancer progression monitoring
3. Epigenetic Analysis Technologies
3.1 DNA Methylation Analysis
Bisulfite Sequencing Protocol
Sample Processing Pipeline:
1. DNA Extraction: Automated extraction from 500 μL whole blood using magnetic bead-based purification (yield: 2-10 μg genomic DNA)
2. Bisulfite Conversion: Treatment with sodium bisulfite converts unmethylated cytosines to uracil while methylated cytosines remain unchanged (conversion efficiency >99%)
3. Library Preparation: Illumina TruSeq DNA Methylation workflow with unique molecular identifiers (UMIs) for error correction
4. Sequencing: NovaSeq 6000 platform generating 100 bp paired-end reads at 30x coverage for comprehensive CpG site analysis
Targeted Methylation Arrays
For routine monitoring, custom Illumina Infinium methylation arrays interrogate 850,000 CpG sites enriched in regulatory regions, focusing on established epigenetic biomarkers for aging (Horvath clock), cancer (methylation-based tumor classification), and metabolic health. Processing time: 48 hours from sample to results.
3.2 Histone Modification Analysis
ChIP-Sequencing for Active Chromatin Marks
Chromatin immunoprecipitation followed by sequencing (ChIP-seq) identifies active regulatory regions:
H3K4me3 (Promoter Activity): Antibody-based pulldown of histone H3 trimethylated at lysine 4, marking active gene promoters
H3K27ac (Active Enhancers): Detection of acetylation at H3 lysine 27, identifying tissue-specific enhancer activity
H3K27me3 (Repressed Regions): Trimethylation marking Polycomb-repressed chromatin, critical for developmental gene regulation
ATAC-Sequencing for Chromatin Accessibility
Assay for Transposase-Accessible Chromatin with sequencing (ATAC-seq) maps open chromatin regions genome-wide using hyperactive Tn5 transposase. This identifies active regulatory elements without requiring antibodies, enabling comprehensive epigenomic profiling from minimal input (500-50,000 cells).
4. Multi-Omic Data Integration Pipeline
4.1 Transcriptomics: RNA Sequencing
Bulk RNA-Sequencing Protocol:
Sample: Peripheral blood mononuclear cells (PBMCs) isolated via density gradient centrifugation (Ficoll-Paque)
RNA Extraction: TRIzol-based extraction followed by DNase treatment and column purification (typical yield: 1-5 μg total RNA from 5 million PBMCs)
Library Construction: Poly-A selection for mRNA enrichment, fragmentation, reverse transcription, and Illumina adapter ligation
Sequencing: 50 million paired-end reads per sample (2×100 bp) on NovaSeq platform, enabling detection of transcripts down to 1 TPM (transcript per million)
Single-Cell RNA-Sequencing (Optional Deep Profiling)
For high-resolution cellular heterogeneity analysis, 10x Genomics Chromium platform captures 5,000-10,000 cells per sample with 3′ or 5′ gene expression profiling. This reveals cell-type-specific transcriptional responses to environmental stressors and disease progression.
4.2 Proteomics: Mass Spectrometry
Plasma Proteomics Workflow
Liquid Chromatography-Mass Spectrometry (LC-MS/MS) Analysis:
Sample Preparation: Immunodepletion of top 14 abundant proteins (albumin, IgG, etc.) to enhance dynamic range, followed by tryptic digestion
Separation: Nanoflow HPLC with C18 reverse-phase column (75 μm ID, 25 cm length) providing high-resolution separation
Detection: Orbitrap Fusion Lumos mass spectrometer with high-field asymmetric waveform ion mobility spectrometry (FAIMS) for enhanced sensitivity
Coverage: Quantification of 3,000-5,000 proteins per sample with coefficient of variation <15% for technical replicates
Targeted Proteomics: SRM/MRM Assays
Selected Reaction Monitoring (SRM) and Multiple Reaction Monitoring (MRM) assays quantify specific protein panels with high precision and reproducibility. Custom assays developed for cardiovascular (troponin, BNP, CRP), metabolic (insulin, leptin, adiponectin), and inflammatory (cytokine panel) biomarkers.

4.3 Metabolomics: Comprehensive Metabolite Profiling
Untargeted Metabolomics
Platform: Combined GC-MS and LC-MS/MS analysis
GC-MS: Detection of volatile and derivatized metabolites (organic acids, amino acids, sugars) after methoximation-silylation chemistry
LC-MS/MS: Comprehensive profiling of polar and nonpolar metabolites using HILIC and reverse-phase chromatography with positive/negative ionization modes
Detection: >1,000 metabolites identified and quantified, including amino acids, lipids, nucleotides, vitamins, and secondary metabolites
Targeted Metabolomics Panels
Biocrates AbsoluteIDQ p400 HR Kit: Quantification of 408 metabolites (amino acids, biogenic amines, acylcarnitines, sphingolipids, glycerophospholipids)
Custom Targeted Panels: Disease-specific metabolite signatures for diabetes (glucose metabolites, branched-chain amino acids), cardiovascular disease (oxidized lipids, trimethylamine N-oxide), and neurodegenerative conditions (neurotransmitter metabolites)
5. Genomic Analysis & Polygenic Risk Scores
5.1 Whole Genome Sequencing
Platform: Illumina NovaSeq X Plus with PCR-free library preparation
Coverage: 30x mean depth across the genome (>95% of bases covered at ≥20x)
Variant Calling: GATK4 best practices pipeline with joint genotyping, achieving >99.9% accuracy for SNP detection
Annotation: Functional annotation using ANNOVAR, VEP, and ClinVar for clinical variant interpretation
5.2 Polygenic Risk Score Calculation
PRS Computation Framework:
1. Reference GWAS Data: Integration of large-scale genome-wide association studies from UK Biobank, FinnGen, and other consortia (>1 million individuals)
2. PRS Calculation: Weighted sum of risk alleles using LDpred2 or PRS-CS for LD-informed polygenic prediction
3. Risk Stratification: Percentile ranking against population distribution with ancestry-matched reference panels
4. Clinical Integration: PRS combined with family history, clinical risk factors, and biomarkers for comprehensive risk assessment
Disease-Specific PRS Panels
Cardiovascular: Coronary artery disease (CAD), atrial fibrillation, stroke, hypertension (variants from >300 loci)
Metabolic: Type 2 diabetes, obesity, dyslipidemia, non-alcoholic fatty liver disease
Cancer: Breast, prostate, colorectal, lung cancer risk (integrating >200 established risk loci per cancer type)
Neurological: Alzheimer’s disease, Parkinson’s disease, multiple sclerosis
5.3 Pharmacogenomics Integration
Genotyping of pharmacogenomic variants (CYP2D6, CYP2C19, CYP3A4/5, SLCO1B1, VKORC1, etc.) enables personalized medication selection and dosing recommendations following CPIC (Clinical Pharmacogenetics Implementation Consortium) guidelines. This prevents adverse drug reactions and optimizes therapeutic efficacy.
6. AI Architecture & Computational Infrastructure
6.1 Deep Learning Models for Multi-Omic Integration
Transformer-Based Architecture
Model Architecture:
Foundation Model: Custom Transformer architecture with 24 layers, 1024 hidden dimensions, 16 attention heads
Pre-training: Self-supervised learning on >10 billion molecular sequences (genomic, transcriptomic, proteomic) from public databases
Fine-tuning: Task-specific adaptation for disease prediction, biomarker discovery, and risk stratification using clinical cohorts
Multi-Modal Fusion: Cross-attention mechanisms integrate information across genomic, epigenomic, transcriptomic, proteomic, and metabolomic modalities

Causal Inference Framework
Rather than simple correlation analysis, the system employs causal discovery algorithms (PC algorithm, GES algorithm, LiNGAM for linear non-Gaussian models) to construct directed acyclic graphs (DAGs) representing causal relationships between molecular features and disease outcomes. This enables mechanistic understanding and identification of intervention targets.
6.2 Digital Twin Simulation Engine
Agent-Based Modeling
The Digital Epigenetic Twin employs agent-based modeling (ABM) to simulate cellular behavior:
Cell Agents: Individual cells represented as computational agents with gene expression states, metabolic profiles, and epigenetic configurations
Environmental Inputs: Diet, exercise, stress, pollutant exposure, circadian rhythm, and medication effects modeled as environmental signals
Interaction Rules: Gene regulatory networks, metabolic pathways, and cell-cell signaling implemented based on established biological knowledge
Temporal Dynamics: Stochastic differential equations model time-evolution of molecular states with parameter inference from longitudinal patient data
Monte Carlo Simulation for Risk Prediction
The Digital Twin runs 10,000+ Monte Carlo simulations per individual, sampling parameter uncertainty and environmental variability to generate probabilistic disease risk trajectories. This produces confidence intervals for predictions and identifies critical intervention windows where preventive actions have maximum impact.
6.3 Computational Hardware Infrastructure
Cloud Architecture:
Primary Platform: Google Cloud Platform with TPU v4 Pods for Transformer model training and inference
Storage: Multi-petabyte BigQuery data warehouse for structured omics data with columnar storage optimization
Genomic Processing: GATK-based pipelines running on Compute Engine with local SSD for high I/O operations
Real-time Processing: Apache Kafka streams for wearable sensor data ingestion (100,000+ events/second throughput)
Security: Data encrypted at rest (AES-256) and in transit (TLS 1.3), with blockchain-anchored audit trails for genetic data access
7. Environmental & Population Sensing Infrastructure
7.1 Nano-Biosensor Network Specifications
Airborne Pathogen Detection
CRISPR-Based Biosensor Technology:
Platform: SHERLOCK (Specific High-sensitivity Enzymatic Reporter unLOCKing) system for nucleic acid detection
Detection Mechanism: Cas13a enzyme coupled with fluorescent reporters provides visual readout within 60 minutes
Sensitivity: Attomolar (10^-18 M) detection limits enabling single-copy pathogen identification
Deployment: Autonomous sampling stations in public spaces (airports, schools, workplaces) with 4-hour sampling cycles and automated cloud reporting
Water Quality Monitoring
Ion-Selective Electrodes: Real-time monitoring of heavy metals (Pb²⁺, Cd²⁺, Hg²⁺), nitrates, and fluoride with ppb-level detection
Microbial Detection: ATP bioluminescence and qPCR-based bacterial/viral quantification with 4-hour turnaround
Chemical Sensors: Gas chromatography-based analysis of volatile organic compounds (VOCs) and endocrine disrupting chemicals
7.2 Population-Level Anomaly Detection
Federated Learning Architecture
Privacy-preserving machine learning where models train locally on user devices and only model updates (not raw data) are shared with central servers. Differential privacy guarantees (ε = 1.0) ensure individual data cannot be reverse-engineered from aggregate statistics.
Outbreak Detection Algorithm
Statistical Approach:
Baseline Modeling: Bayesian hierarchical models establish expected biomarker distributions for each geographic region and time period
Anomaly Scoring: Mahalanobis distance calculation identifies multivariate deviations from baseline (>3 standard deviations trigger alerts)
Spatial Clustering: SaTScan software detects geographic clusters of elevated biomarkers using space-time permutation scans
Validation: Automated alerts verified through environmental sensor data and healthcare system integration (emergency department syndromic surveillance)
8. Clinical Validation & Regulatory Strategy
8.1 Clinical Study Design
Prospective Cohort Study
Study Parameters:
Population: 50,000 participants aged 40-75 with diverse ancestries, stratified by baseline disease risk
Follow-up: 5-year longitudinal tracking with quarterly biomarker assessments and annual comprehensive omics profiling
Endpoints: Incident cardiovascular events, cancer diagnoses, metabolic disease progression, all-cause mortality
Statistical Power: 90% power to detect hazard ratio of 1.3 for primary endpoints at α = 0.05 with adjustment for multiple testing
Intervention Trial
Randomized controlled trial comparing Digital Twin-guided preventive interventions versus standard care. Primary outcome: reduction in composite cardiovascular events at 3 years. Secondary outcomes: quality of life, healthcare utilization, and cost-effectiveness.
8.2 Regulatory Pathway
FDA Regulatory Strategy
Device Classification: Class II medical device (Software as a Medical Device – SaMD) under FDA Digital Health guidance
Pre-Cert Program: Participation in FDA Pre-Certification pilot for expedited review based on organizational excellence
Clinical Evidence: Submission of analytical validation, clinical validation, and clinical utility evidence from prospective studies
CLIA Certification: Laboratory operations certified under Clinical Laboratory Improvement Amendments for diagnostic testing
International Approvals
CE Mark (Europe): In vitro diagnostic medical device under IVDR regulations with notified body assessment
PMDA (Japan): Collaboration with PMDA for approval under regenerative medicine regulations
NMPA (China): Phased approval strategy beginning with Class II approval for genomic analysis components
9. Data Security & Privacy Architecture
9.1 Blockchain-Based Genetic Data Storage
Implementation:
Platform: Hyperledger Fabric permissioned blockchain with Practical Byzantine Fault Tolerance consensus
Data Structure: Genetic data encrypted with user-specific keys; blockchain stores hashes and access logs for immutable audit trail
Smart Contracts: Automated consent management and data access permissions enforced by chaincode
User Control: Granular permissions allowing users to approve/revoke data access for specific purposes or researchers
9.2 Differential Privacy Implementation
All population-level statistics computed using differential privacy with calibrated Laplace noise addition (privacy budget ε = 1.0). This provides mathematically rigorous privacy guarantees while enabling accurate aggregate analysis for outbreak detection and health trends.
9.3 Compliance Framework
HIPAA: Full compliance with Security Rule (access controls, encryption, audit logging) and Privacy Rule (minimum necessary, consent management)
GDPR: Data minimization, right to erasure, data portability, and consent management for EU users
GINA: Compliance with Genetic Information Nondiscrimination Act protecting against insurance/employment discrimination
SOC 2 Type II: Annual attestation of security controls by independent auditors
10. Cost Structure & Economic Model
10.1 Per-User Cost Breakdown

| Component | Initial Cost | Annual Recurring |
| Whole Genome Sequencing (30x) | $300 | — |
| Epigenetic Array (baseline) | $150 | $150 (annual) |
| RNA-Seq (quarterly) | — | $400 (4x $100) |
| Proteomics (semi-annual) | — | $600 (2x $300) |
| Metabolomics (quarterly) | — | $200 (4x $50) |
| Wearable Biosensor Device | $200 | $120 (sensor refills) |
| AI/Cloud Infrastructure | — | $180 |
| Total | $650 | $1,650/year |
10.2 Cost Reduction Strategies
AI-Powered Imputation: Predicting full genomic profiles from low-pass sequencing (1x coverage) reduces initial sequencing costs by 75% while maintaining >98% accuracy for common variants
Batch Processing: Multiplexed sample processing reduces reagent costs by 40% compared to individual assays
Automated Workflows: Robotic liquid handling and automated library prep reduce labor costs by 60%
Scale Economics: At 1 million+ users, sequencing costs projected to decrease to <$100/genome through vertical integration and strategic partnerships
Technical Summary
The NEXUS-GENE platform represents a fully integrated, technically feasible system for real-time health monitoring and population sensing. Every component—from electrochemical biosensors to transformer-based AI models—builds on established, validated technologies. The innovation lies in the comprehensive integration of multi-omic profiling, continuous wearable monitoring, environmental sensing, and causal AI into a unified anticipatory health system.
With clear clinical validation pathways, regulatory strategies, and economic models demonstrating viability at scale, NEXUS-GENE is positioned to transform healthcare from reactive treatment to proactive prevention. The technical infrastructure described in this document provides the foundation for a new era of personalized, predictive, and participatory medicine.
Legal Notice & Intellectual Property Rights
© 2026 Dr. Dimitris Kimoglou | Bravanton Institute. All Rights Reserved.
This text and the NEXUS-GENE framework are the exclusive intellectual property of Dr. Dimitris Kimoglou and the Bravanton Institute. This publication is intended solely for informational purposes regarding the official submission of the proposal to MIT Solve.
Any partial or total reproduction, copying, republication, or unauthorized use of the technical terminology, algorithmic descriptions, and scientific methodologies contained herein is strictly prohibited without prior written consent from the author. The underlying concept and technical architecture are protected under international intellectual property laws. Unauthorized use or infringement will result in legal action.










